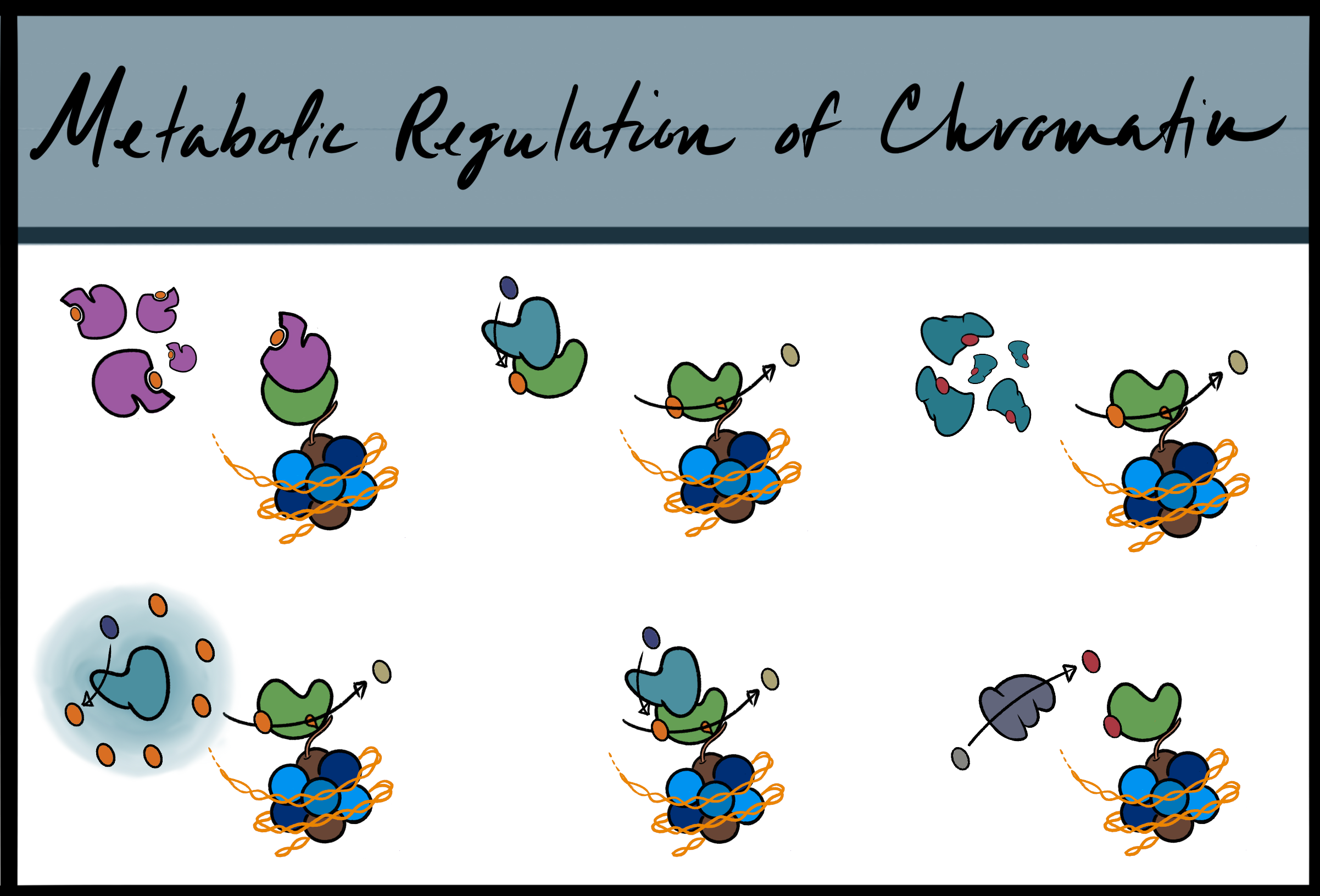
RESEARCHRepresentative images of the diverse mechanisms of metabolic proteins in regulating chromatin-assocaited proteins.
METABOLIC REGULATION OF CHROMATIN
While metabolism is primarily thought of as a network of biochemical reactions that maintain nutrient availability for energy production, metabolites from these reactions also serve as substrates and cofactors for epigenetics enzymes. In fact, an increasing number of metabolic proteins have been found to translocate into the nucleus and regulate chromatin functions upon internal and external cues. Our lab focuses on understanding this metabolic-epigenetic relationship in the context of aging and how we can target this axis to promote healthy aging.
RELEVANT PUBLICATIONS
Tran KA, Dillingham CM, Sridharan R. Coordinated removal of repressive epigenetic modifications during induced reversal of cell identity. EMBO J. 2019 Nov 15;38(22):e101681. PubMed Central PMCID: PMC6856600.
Tran KA, Dillingham CM, Sridharan R. The role of α-ketoglutarate-dependent proteins in pluripotency acquisition and maintenance. J Biol Chem. 2019 Apr 5;294(14):5408-5419. PubMed Central PMCID: PMC6462505.
Tran KA, Jackson SA, Olufs ZP, Zaidan NZ, Leng N, Kendziorski C, Roy S, Sridharan R. Collaborative rewiring of the pluripotency network by chromatin and signaling modulating pathways. Nat Commun. 2015 Feb 4;6:6188. PubMed Central PMCID: PMC4347202.
Image demonstrating the dysregulation of chromatin proteins (epigenetic enzymes, cofactors, and transcription factors, etc..) during aging leading to altered gene expression.
CHROMATIN DYNAMICS DURING AGING
During senescence and aging, there is specific and global changes in chromatin architecture leading to altered gene expression. In fact, targeting epigenetic regulators has been shown to promote longevity and ameliorate age-related disorders. Our lab investigates how chromatin dynamics is misregulated during aging using various epi-genomics approaches. We hope to find mechanisms to perserve chromatin integrity to promote healthy aging.
RELEVANT PUBLICATIONS
Wang L, Donahue G, Zhang C, Havas A, Lei X, Xu C, Wang W, Vahedi G, Adams PD, Berger SL. Dynamic enhancer interactome promotes senescence and aging. bioRxiv [Preprint]. 2023 May 22:2023.05.22.541769. doi: 10.1101/2023.05.22.541769. PMID: 37292952; PMCID: PMC10245931.
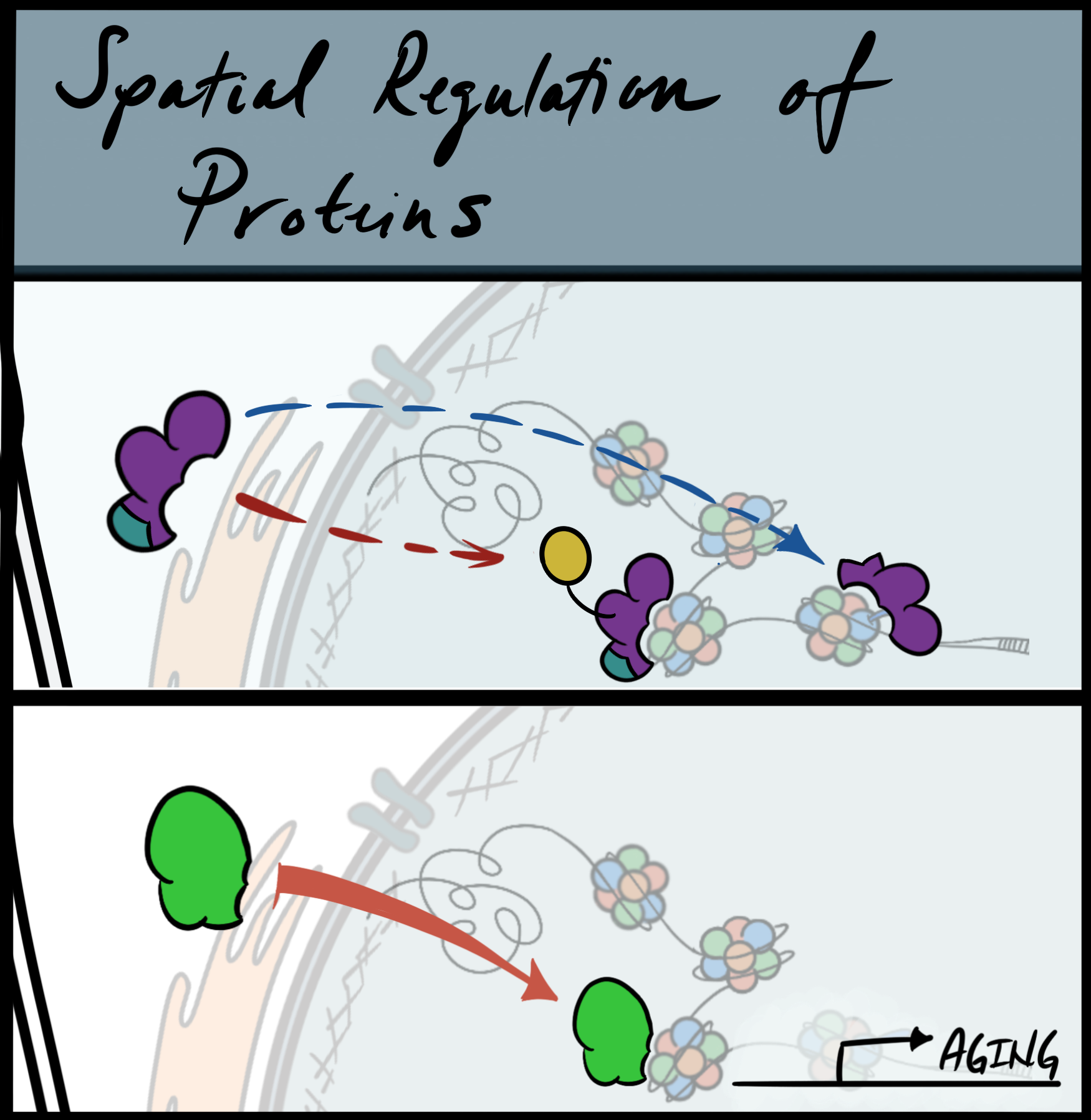
(Top) Representative depiction of possible mechanisms regulating spatial localization of proteins (isoform switching or post-translation modifications). (Bottom) representation of how protein localization is altered during aging.
SPATIAL REGULATION OF PROTEINS
Proteins are highly dynamic and their misregulation can lead to cellular dysfunction and disease. Using a conjunction of imaging and proteomics approaches, our lab studies how protein localization is govern as spatial regulation can influence protein environment, protein-protein interactions, post-translation modifications and substrate binding. The appropriate coordination of protein homeostasis is critical for healthy aging as dysregulation of this process can lead to accelerated aging and age-related disorders.
RELEVANT PUBLICATIONS
Tran KA, Gilbert M, Vazques BN, Ianni A, Garcia BA, Vaquero A, Berger SL. SIRT7 regulates NUCKS1 chromatin binding to elicit metabolic and inflammatory gene expression in senescence and liver aging. Mol Cell. 2025 Jun 19;85(12):2390-2408.e6. PubMed Central PMCID: PMC12225685
Yu R. Roseman S, Siegenfeld AP, Gardner Z, Nguyen SC, Tran KA, Joyce EF, Jain R, Liau BB, Krantz ID, Alexander KA, Berger SL. CTCF/RAD21 organize the ground state of chromatin-nuclear speckle association. Nat Struct Mol Biol. 2025 Jun;32(6):1069-1080. PubMed Central PMCID: PMC12170152